Effects of serum matrix on the molecular interactions between drugs and target proteins as revealed by GMR biosensor technology
Investigations of molecular interactions between drugs and biomolecules are indispensable for understanding the mechanism of drug actions and their pharmacokinetics/pharmacodynamics. However, analyses of protein interactions are typically performed in buffered salt solutions which lack many components that are present in the original biological matrix, and thus has significant effects on the binding behaviors observed. This limitation of not being able to conduct binding kinetic studies in relevant biological matrices is a result of using an inadequate technology. The standard technology for such studies is based on surface plasmon resonance (SPR) measurements where nonspecific binding due to blood proteins distorts the kinetic data for samples with more than 1% serum. Therefore, detailed kinetic studies in relevant biological matrices are unavailable.
In a recent study published in J. Pharm. Biomed. Anal. 198, 114015 (2021), Saito, et al., used the Giant Magneto Resistance (GMR) platform from MagArray to investigate the binding characteristics of three molecular pairs in a serum-based matrix compared with a buffer-only matrix. The pairs were: quercetin and cAMP-dependent protein kinase A (PKA), Infliximab and tumor necrosis factor alpha (TNFα), and Bevacizumab and vascular endothelial growth factor (VEGF). They found that the presence of serum had different effects on the binding kinetics of the pairs compared to buffer-only measurements. Protein pairs were not as affected as were the small molecules. Saito et al., attributed the differences to the hydrophobic and electrostatic characteristics of the drug molecule, target protein, and serum proteins, which could only be evaluated with the GMR technology. They concluded that the real-time monitoring of molecular interactions in a more relevant biological matrix, enabled by the GMR platform, was a powerful tool to investigate such complicated, yet real-life, molecular interactions. Additionally, the method would allow the analysis of any kind of effects by a third molecule on the interaction between two other molecules, for example, an inhibitor drug’s interactions between two kinds of proteins. Such capabilities expand the depth of drug discovery and characterization.
To learn more about how the MagArray GMR platform can also help your drug discovery and characterization efforts, click here, or contact Kalidip Choudhury, Ph.D., at MagArray Lifesciences kalidip.choudhury@magarray.com
Giant Magnetic Resonance (GMR) Assay Principles
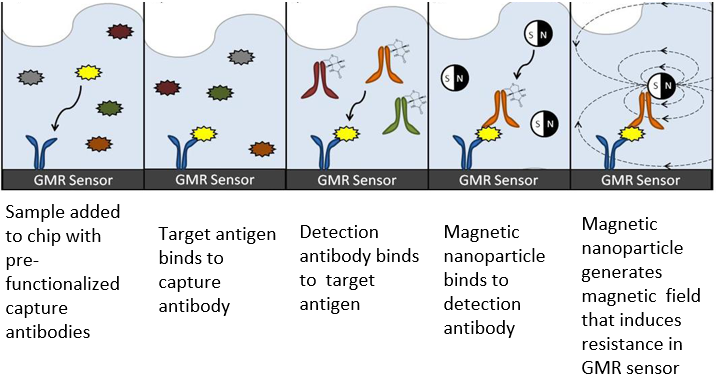
Our platform utilizes a sandwich assay in which the target antigen is sandwiched between two antigens, one bound to the magnetic sensor and the other to a superparamagnetic nano particle. Under an external magnetic field, the nanoparticles magnetize and their presence or absence can be detected by the sensor. Using chips with 64 GMR sensors we show rapid multiplex protein detection with a liner dynamic range of over 5-6 orders of magnitude for a diverse range of biological fluids in real time.
*Based on standard LBA process with magnetic nanoparticles used as labels

Instrument and chip
GMR Multiplex Chip
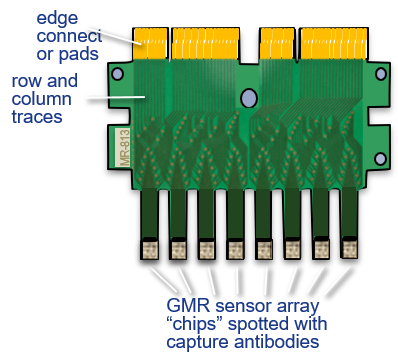
- 80 individual sensors on each chip
- 8 chips with 640 sensors total
- Each sensor can be individually spotted
- 80 biomarkers with controls for the multiplex chip
GMR Reader Unit
- Rugged and reliable
- No optics meaning no calibration of lasers, mirrors etc
- No fluidics means no pumps, valves, etc
- Fully automated
- Hands free incubation
- Bar code system to minimize errors
- No wash necessary
- Flexible assay protocols
- Run time of 1 hour for most sensitive assay
- Run time of 5-10 minutes for express assays
Multiplex Immunoassay
MagArray has 5-6 orders of magnitude greater dynamic range and a 1000 times more sensitive than ELISA.
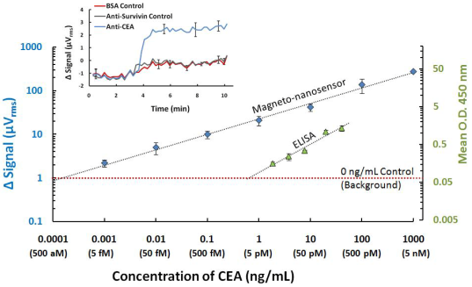
Compared to ELISA, the MagArray platform is capable of detecting CEA at significantly lower concentrations and at 5-6 orders of magnitude greater dynamic range utilizing the same Ab pair
Matrix insensitive detection in different buffers.
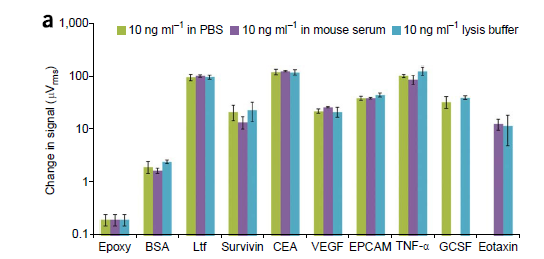
A panel of eight human tumor markers with BSA negative control and epoxy control indicates matrix insensitive detection when shifting from PBS to mouse serum to lysis buffer
Ligand Binding Assay
Our GMR platform is more sensitive than SPR and insensitive to buffer conditions
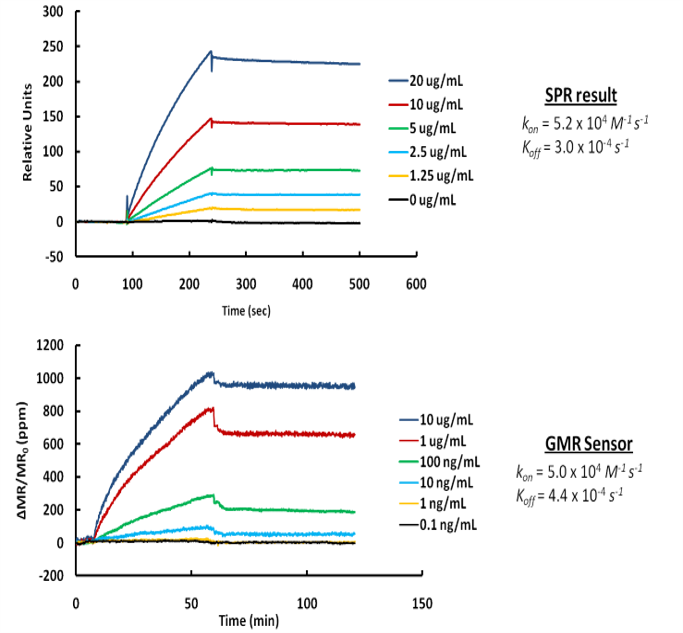
MagArray platform at least 1000 times more sensitive than Biacore/SPR
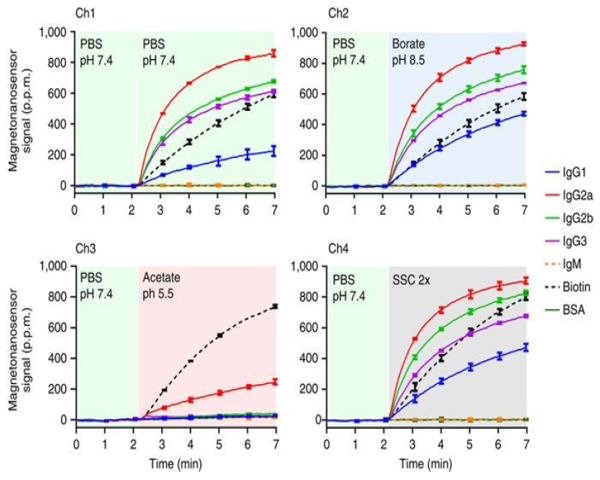
MagArray platform insensitive to pH and salinity*
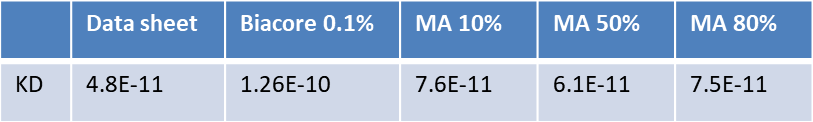
The effect of matrix on protein binding between SPR and MagArray
Buffer: 0.01M HEPES, pH 7.4, 0.15M NaCl, 3mM EDTA
Regeneration: 10mM Gly-HCL, pH 2.5
Analyte: TSH, Ligand: Anti-TSH ab
Biacore X 100
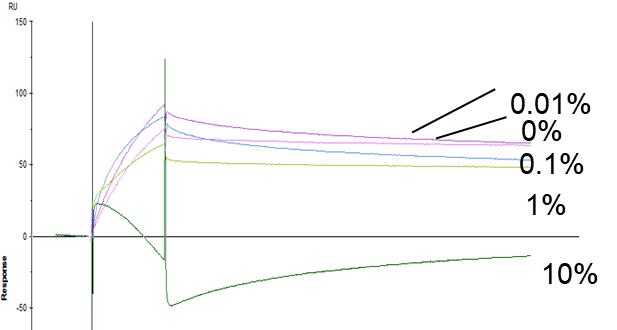
- The same binding pair and buffer conditions were used to compare between MagArray and Biacore
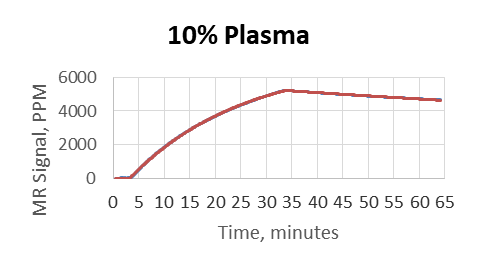
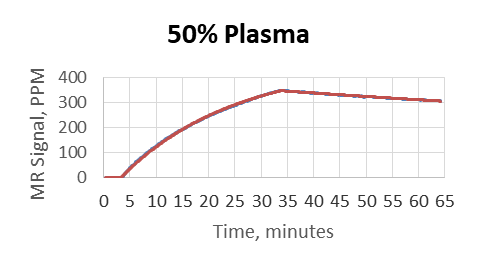
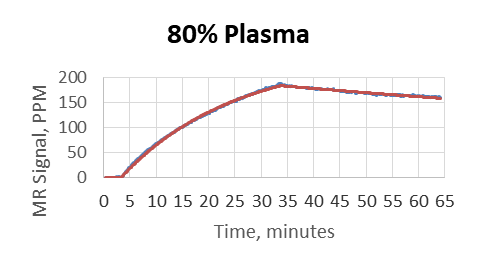
- Plasma was used as a matrix and diluted by PBS to 80%, 50%, 10%, 1%, 0.1%
- MagArray platform kinetic measurements were not affected by matrix
- Biacore did not provide reliable results above 0.1% matrix
MagArray
- Truly a transformational platform that helps to bridge the gap between in-vitro and in-vivo.
- Matrix insensitive technology for measuring biomolecules in blood, plasma, serum, saliva and urine.
- 1000 times more sensitive than leading SPR technologies in measuring protein-protein interactions.
- Partnership with Hitachi Hi-Technologies allows expansion of platform for unmet needs in biological assays.
Publications
- R. S. Gaster, D. A. Hall, C. H. Nielsen, S. J. Osterfeld, H. Yu, K. E. Mach, R. J. Wilson, B. Murmann, J. C. Liao, S. S. Gambhir & S. X. Wang, “Matrix-insensitive protein assays push the limits of biosensors in medicine,” Nature Medicine, Oct. 11 (online publication), 2009.
Vermesh O, Aalipour A, T. Ge J, Saenz Y, Guo Y, Alam IS, Park S-min, Adelson CN, Mitsutake Y, Vilches-Moure J et al.. 2018. An intravascular magnetic wire for the high-throughput retrieval of circulating tumour cells in vivo. Nature Biomedical Engineering. 2:696-705.Google Scholar
Lee J.R., Appelmann I, Miething C, Shultz T.O., Ruderman D, Kim D, Mallick P, Lowe S.W., Wang S.X.. 2018. Longitudinal Multiplexed Measurement of Quantitative Proteomic Signatures in Mouse Lymphoma Models Using Magneto-Nanosensors. Theranostics. 8(5)Google Scholar
Ng E, Yao C, Shultz TO, Ross-Howe S, Wang SX. 2018. Magneto-nanosensor Smartphone Platform for the Detection of HIV and Leukocytosis at Point-of-Care. Nanomedicine: Nanotechnology, Biology and Medicine. Google Scholar
Ravi N, Rizzi G, Chang SE, Cheung P, Utz PJ, Wang SX. 2018. Quantification of cDNA on GMR biosensor array towards point-of-care gene expression analysis. Biosensors and Bioelectronics. Google Scholar
Beggs M., Yu H., Carbonell L., Wang S.X., Vachani A.. 2018. Validation of Plasma TIMP-1 to Identify Lung Cancer in Smokers. D99. CLINICALLY INFORMATIVE BIOMARKERS IN LUNG CANCER: A NEEDLE IN A HAYSTACK. :A7415-A7415.Google Scholar
Lee J-R, Chan CT, Ruderman D, Chuang H-Y, Gaster RS, Atallah M, Mallick P, Lowe SW, Gambhir SS, Wang SX. 2017. Longitudinal Monitoring of Antibody Responses against Tumor Cells Using Magneto-nanosensors with a Nanoliter of Blood. Nano Letters. 17:6644-6652.Google Scholar
Park S-min, Wong DJ, Ooi CChun, Nesvet JC, Nair VS, Wang SX, Gambhir SS. 2017. Multigene profiling of single circulating tumor cells.Molecular & Cellular Oncology. 4:e1289295.Google Scholar
Rizzi G, Lee J-R, Dahl C, Guldberg P, Dufva M, Wang SX, Hansen MF. 2017. Simultaneous Profiling of DNA Mutation and Methylation by Melting Analysis Using Magnetoresistive Biosensor Array. ACS Nano. 11:8864-8870.Google Scholar
Ng E, Nadeau KC, Wang SX. 2016. Giant magnetoresistive sensor array for sensitive and specific multiplexed food allergen detection.. Biosens Bioelectron. 80:359-365.PubMed
Lee J-R, D Haddon J, Gupta N, Price JV, Credo GM, Diep VK, Kim K, Hall DA, Baechler EC, Petri M et al.. 2016. High-Resolution Analysis of Antibodies to Post-Translational Modifications Using Peptide Nanosensor Microarrays.. ACS Nano. PubMed
Brett E, Zielins ER, Luan A, Ooi CChun, Shailendra S, Atashroo D, Menon S, Blackshear C, Flacco J, Quarto N et al.. 2016. Magnetic Nanoparticle‐Based Upregulation of B‐Cell Lymphoma 2 Enhances Bone Regeneration. STEM CELLS Translational Medicine. 6:151-160.Google Scholar
Lee J-R, Bechstein DJB, Ooi CChun, Patel A, Gaster RS, Ng E, Gonzalez LC, Wang SX. 2016. Magneto-nanosensor platform for probing low-affinity protein-protein interactions and identification of a low-affinity PD-L1/PD-L2 interaction.. Nat Commun. 7:12220.PubMed
Park S-min, Wong DJ, Ooi CChun, Kurtz DM, Vermesh O, Aalipour A, Suh S, Pian KL, Chabon JJ, Lee SHun et al.. 2016. Molecular profiling of single circulating tumor cells from lung cancer patients. Proceedings of the National Academy of Sciences. 113:E8379-E8386.Google Scholar
Lee J-R, D Haddon J, Wand HE, Price JV, Diep VK, Hall DA, Petri M, Baechler EC, Balboni IM, Utz PJ et al.. 2016. Multiplex giant magnetoresistive biosensor microarrays identify interferon-associated autoantibodies in systemic lupus erythematosus.. Sci Rep. 6:27623.PubMed
Choi J, Gani AWijaya, Bechstein DJB, Lee J-R, Utz PJ, Wang SX. 2016. Portable, one-step, and rapid GMR biosensor platform with smartphone interface.. Biosens Bioelectron. 85:1-7.PubMed
Lee J-R, Choi J, Shultz TO, Wang SX. 2016. Small Molecule Detection in Saliva Facilitates Portable Tests of Marijuana Abuse.. Anal Chem. 88(15):7457-61.PubMed
Earhart CM, Hughes CE, Gaster RS, Ooi CChun, Wilson RJ, Zhou LY, Humke EW, Xu L, Wong DJ, Willingham SB et al.. 2014. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips.. Lab Chip. 14(1):78-88.PubMed
Partner With Us
For pharmaceutical life science inquiries email Kalidip.Choudhury@magarray.com.
We are always looking for new applications for our technology. Talk with us about how we might help you.